Human Genome Project: An Introduction
The Human Genome Project is based on the fact that isolating and analysing the genetic material contained in DNA can provide scientists with powerful new approaches to understanding disease development and developing new strategies for disease prevention and treatment. Except for physical injuries, nearly all human medical conditions are linked to changes (i.e., mutations) in the structure and function of DNA. The HGP accelerated the growth of bioinformatics, a vast field of study.
The project's primary goal is to create research tools that enable scientists to identify genes involved in rare and common diseases. In this article, we will study the various features of this megaproject as well as its applications in various fields, and the steps taken up by scientists to sequence the whole genome.
What is the Human Genome Project?
The Human Genome Project is an international research project with the primary goal of deciphering the chemical sequence of the entire human genetic material (i.e., the entire genome). It identifies all 50,000 to 100,000 genes contained within the genome and provides research tools to analyse all of this genetic information.
After the US government picked up the idea in 1984 and began planning, the project was formally launched in 1990 and completed in 2003.
The National Institutes of Health (NIH) of the United States, as well as numerous other organisations from around the world, provided funding.
The Human Genome Project (HGP) aims to determine the sequence of chemical base pairs that comprise human DNA, map the entire human genome, and identify its complex structures and functions.
Differences in the genetic make-up are caused by differences in DNA nucleotide sequences. The goal of scientists has always been to map the human genome. Advances in genetic engineering techniques have made it possible to isolate and clone DNA fragments and determine their nucleotide sequences.
The HGP has transformed biology with its multidisciplinary approach to deciphering a reference human genome sequence.
This audacious endeavour resulted in the creation of novel technologies and analytical tools.
Finally, the HGP has inspired several other exciting projects that have the potential to open up new avenues in biology, medicine, and psychology.
Aim and Objective of the Human Genome Project
To sequence the whole genome at 3 billion bps.
To create a physical map of the human genome.
To store this information in the database.
To improve the tools for data analysis.
To transfer this information to the other related industries.
To solve any ethical, legal, or social issues regarding this project.
To make the information available to all the researchers.
Steps of the Human Genome Project
The whole DNA of the cell is isolated and randomly broken into fragments.
They are inserted into special vectors like BAC (Bacterial Artificial Chromosomes) and YAC (Yeast Artificial Chromosomes).
These fragments are then cloned into suitable hosts like bacteria and yeast.
A Polymerase Chain Reaction (PCR) is used to make copies of DNA fragments.
The fragments are sequenced using Sanger sequencing.
The sequences are then arranged based on the overlapping regions.
The sequences were then annotated and assigned to different chromosomes.
The genetic and physical maps are also made with the help of polymorphism of microsatellites and restriction endonuclease.

Steps used in Human Genome Project Image
Salient Features of the Human Genome Project
The human genome is made up of 3164.7 million nucleotides.
The average gene is 3000 base pairs long. On the X-chromosome, the largest gene is Duchenne Muscular Dystrophy. It has 2.4 million base pairs (2400 kilo). The genes for B-globin and insulin are less than 10 kilobases long.
The human genome contains approximately 30,000 genes. It was previously estimated that it contained 80,000 to 100,000 genes. The number of genes in humans is roughly equal to that of mice.
More than half of the discovered genes' functions are unknown.
Proteins are coded for in less than 2% of the genome.
Repetitive sequences are nucleotide sequences that are repeated hundreds or thousands of times. They do not directly code but provide information about chromosome structure, dynamics, and evolution.
Approximately 1 million copies of short 5-8 base pair repeated sequences are clustered around centromeres and near the ends of chromosomes. They represent junk DNA.
Chromosome I has the most genes (2968) and Y has the fewest (231).
In humans, there are approximately 1.4 million locations where single-base DNA differences (SNPs- Single nucleotide polymorphism) occur.
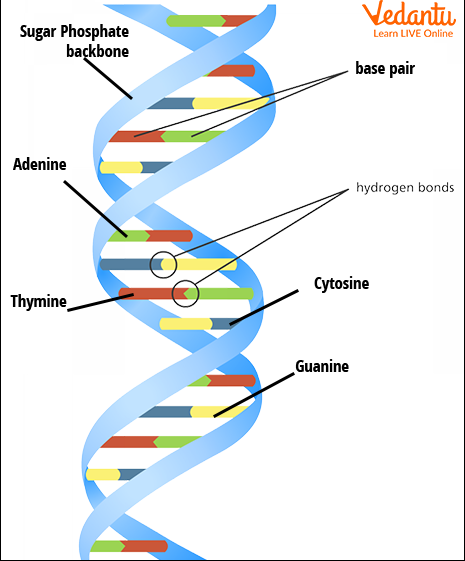
Structure of DNA
The Technique Used in HGP:
The Human Genome Project used Sanger sequencing to determine the sequences of relatively small fragments of human DNA (900 bp or less).
These fragments were then used to piece together larger DNA fragments and, eventually, entire chromosomes.
The advancement of next-generation sequencing (NGS) technologies has accelerated genomics research.
Applications of Human Genome Project
Gene discovery also opens up the possibility of developing gene-based treatments for both hereditary and acquired diseases.
It's detailed genetic, physical, and sequence maps will also be critical in understanding the biological basis of complex disorders caused by the interaction of multiple genetic and environmental influences, such as diabetes, heart disease, cancer, and psychiatric illnesses such as alcoholism.
It helps in the identification of mutations linked to different forms of cancer.
It also helps in advancing research in Forensic Sciences.
Agriculture, environment, and biotechnology are some other fields that have benefitted from the use of human genome projects.

Diversity of Genomic Applications to Various Fields
Conclusion
The Human Genome Project (HGP) is an international scientific research project that aimed to identify, map, and sequence all of the genes in the human genome from both a physical and functional standpoint. Each individual's "genome" is unique; mapping the "human genome" requires sequencing a small number of individuals and then assembling these to obtain a complete sequence for each chromosome. As a result, the completed human genome is a combination that does not represent any single individual.


FAQs on Human Genome Project
1. What exactly was the Human Genome Project (HGP)?
The Human Genome Project was a landmark international research effort that successfully determined the entire sequence of nucleotide base pairs that make up human DNA. Launched in 1990 and completed in 2003, its primary goal was to identify and map all of the genes of the human genome from both a physical and functional standpoint.
2. What is a 'genome' in simple terms?
A genome is the complete set of genetic instructions for an organism. It contains all the information, stored in its DNA, that is needed for the organism to build and maintain itself. Think of it as the master blueprint for all cellular structures and activities for the entire lifespan of the cell or organism.
3. What were the main goals of the Human Genome Project?
The HGP had several key objectives as per the CBSE/NCERT curriculum for the 2025-26 session. The major goals were to:
- Identify all the approximately 20,000-25,000 genes in human DNA.
- Determine the sequences of the 3 billion chemical base pairs that make up human DNA.
- Store this vast amount of information in public databases.
- Develop faster, more efficient tools for data analysis.
- Address the Ethical, Legal, and Social Issues (ELSI) that might arise from the project.
4. What is the difference between the two main sequencing methods used in the HGP?
The two major approaches were Expressed Sequence Tags (ESTs) and Sequence Annotation. ESTs involved identifying only the genes that are expressed as RNA, providing a faster but incomplete picture. Sequence Annotation was a more comprehensive method where the entire genome was sequenced first, and then the coding and non-coding regions were identified and assigned functions.
5. What are BAC and YAC, and why were they important for the HGP?
BAC stands for Bacterial Artificial Chromosome and YAC stands for Yeast Artificial Chromosome. These are vectors used in genetic engineering to clone large fragments of DNA. They were essential for the HGP because the human genome is too large to sequence in one go. Scientists used BACs and YACs to break the genome into smaller, manageable pieces, which were then cloned and sequenced individually.
6. What are some of the most important applications of the Human Genome Project today?
The knowledge from the HGP has revolutionised many fields. In medicine, it allows for a better diagnosis of diseases, early detection of genetic disorders, and the development of gene therapy and personalised medicine. It has also greatly advanced research in biotechnology and has become a fundamental tool in understanding human evolution and forensics.
7. It is often said that less than 2% of our genome codes for proteins. What does the other 98% do?
This is a fascinating aspect of our genome. The vast majority of our DNA that does not code for proteins is not 'junk'. This non-coding DNA has crucial functions, including regulating gene expression (controlling when and where genes are turned on or off), providing structural integrity to chromosomes, and playing roles in evolution. Scientists are still actively researching the full purpose of this 'dark matter' of the genome.
8. What were some of the most surprising findings from the HGP?
The HGP revealed several unexpected facts about our genetic makeup:
- The human genome contains only about 20,500 protein-coding genes, much fewer than the initial estimate of 100,000.
- All human beings are 99.9% identical at the DNA level.
- The functions of over 50% of the discovered genes are still unknown.
- Chromosome 1 has the most genes (2968), while the Y chromosome has the fewest (231).
9. How does understanding the human genome help in studying genetic disorders?
By having a complete 'map' of the human genome, scientists can now more easily identify the specific genes or mutations responsible for genetic disorders like cystic fibrosis, sickle cell anaemia, and Huntington's disease. This knowledge is the first step towards developing better diagnostic tests, predicting risk, and creating targeted treatments to correct or manage these conditions.
10. What are the 'ELSI' associated with the Human Genome Project?
ELSI stands for Ethical, Legal, and Social Issues. These are concerns about the potential consequences of having detailed genetic information. Key issues include the fair use of genetic information, preventing genetic discrimination by employers or insurance companies, ensuring the privacy and confidentiality of an individual's genetic data, and the psychological impact of knowing one's genetic predispositions.
11. If all humans are 99.9% genetically similar, what accounts for our vast individual differences?
The immense diversity among humans is packed into the tiny 0.1% of our DNA that differs between individuals. This small fraction contains millions of genetic variations, most commonly as Single Nucleotide Polymorphisms (SNPs). These SNPs are single base-pair changes that, along with other variations, are responsible for all our unique traits like eye colour, height, and susceptibility to various diseases.










